My little understanding about CADD..^_^
Bismillahirrahmanirrahim...
As this blog is created mainly to fulfill the special requirement for CADD class, here, I would like to share a little bit about what i had learned for about those past three lectures. To be frankly speaking, being a student with zero knowledge of bioinformatics, me myself could feel that this subject will be a heavy course for this semester. However, when Madam Noraslinda gave this task, I think that this is good way of testing our understanding of this subject besides assist us to understand and revise what she had taught us before. So here is a so called summary of the knowledge which I had gained. As this is merely based on my little understanding, I apology for any mistake..=). Before starting the lesson, in order to make this 'lecture' more understandable, I will divide the 'lecture notes' into subtopics.
Week 1: Bioinformatic tools
Although this course is named as CADD (Computer Aided Drug Design), the basic knowledge that we must know is bioinformatics. Bioinformatics, according to Wikipedia is the application of information technology and computer science to the field of molecular biology. In fact, bioinformatic is the centre where several disciplines are belong to it such as CADD, information technology, application and databases also computational resources. Therefore, in order for us to understand CADD, me must at first being familiar with the bioinformatics tools. In the first class at the computer lab, we were being introduced to the bionformatics tools website and how to utilize those websites. the following is the detail of how to use the website.
MEROPS webpage
This website provides the information resource for peptidases and the proteins that inhibit them. For our class, we were taught of two peptidases, which are ClpP and Lon. Both can be retrieved by clicking at the search button --> search for peptidase..by name --> search for ClpP and Lon --> choose the peptidase S16.001 for Lon-A peptidase and peptidase S14.001 for peptidase Clp (type 1) --> go to "sequence" -- > choose any five organisms for Lon and ClpP each (the MEROPS ID start with MER) --> then paste the sequence in the notepad for the next lecture and our future reference.
After exploring MEROPS, we were taught on how to search the details of the previous chosen organisms by using PSIBLAST from this webpage. After clicking on the "blast" on the right webpage, find for "protein blast".
After clicking at "protein blast", the following will appear and the protein sequence will be entered in the query sequence box at the top on the left of the page.
At the bottom of the page, the Program Selection Algorithm should be changed to PSI-BLAST before go to BLAST button. The detail and further description of the protein sequence would then appeared.
To see the identity and description of the protein, click on the first line to get below feature.
Week 2: ClustalX and Artemis
Basically the second class is still the introduction class in which two new software, ClustalX and Artemis were being introduced.
a) ClustalX
This software is used primarily for loading the sequence available in the notepad file from previous class(Lon and ClpP peptidases). Besides that, other properties such as alignment also be made by using this software.
The loading of Lon and ClpP sequence by using ClustalX
b) Artemis
Artemis is a free genome viewer and annotation tool that allows visualization of sequence features and the results of analyses within the context of the sequence. in other word, it allows the users to choose a specific sequence, for instance Lon and ClpP sequence from Burkholderia pseudomallei to look for its location in the genome. Moreover, we also learned the steps to trim, delete, rename and move the entry of the selected sequences.
c) PDB(Protein Data Bank)
The information from PDB could be found out by knowing the PDB ID. Therefore, by using the same organism, which is Burkholderia pseudomallei, the description or the PDB ID could be identified from the NCBI and BLAST application, as mentioned earlier. Once the PDB ID was found; Lon PDB ID was 1TYF while ClpP PDB ID was 1RR9, the FASTA sequences and PDB file were downloaded.
The information from PDB could be found out by knowing the PDB ID. Therefore, by using the same organism, which is Burkholderia pseudomallei, the description or the PDB ID could be identified from the NCBI and BLAST application, as mentioned earlier. Once the PDB ID was found; Lon PDB ID was 1TYF while ClpP PDB ID was 1RR9, the FASTA sequences and PDB file were downloaded.
Example: This page will be appeared after enter the PDB ID of 1TYF. The further step is to click at download files and download FASTA sequence and PDB file(gz).
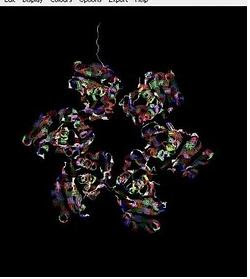
Week 3: Protein 3D formation
Using the information from PDB file, we learned on how to form protein 3D structure with the assistance of a new software; Raswin. It was really amazing to see the protein being formed was in the colorful and real figure.
~I hope that this post will at least give a brief idea on what happen in CADD class..=)
Wassalam..
Read Users' Comments (0)